Neurological Disorders:
Amyotrophic Lateral Sclerosis (ALS),Multiple Sclerosis (MS), Stroke, Traumatic Brain Injury (TBI), Spinal cord Injury (SCI), Brain Tumor
Neuroimaging: Structural MRI, functional MRI, Diffusion MRI, PET,
Electrophysiology: EEG, Transcranial Magnetic Stimulation (TMS), EMG, Functional Electrical Stimulation (FES)
Other Research Areas: Biomechanics, Human Gait Analysis, Machine Learning, Neural Networks, Graph Theory (Brain Network Analysis)
Skills and Expertise: Diffusion MRI single shell, multi shell, Conventional MRI T1-w, T2-w, T2 relaxometry, Voxel Based Morphometry, Tensor Based Spatial statistics, cortical thickness analysis, TMS, EEG, EMG data processing, Tractography, Fractal analysis (mono and multi-fractal), ICA
Neuroimaging Softwares: FSL, SPM, DTI studio, Freesurfer, DSI studio, ExploreDTI, GAT, EEG Lab
Neuroimaging Research in Amyotrophic Lateral Sclerosis (ALS)
Understanding brain network in ALS phenotypes a) ALS patients with corticospinal tract hyperintensity in T2/Pd-W images, b) ALS patients without CST hyperintensity in T2/PD-W images, c) Classic ALS, d) ALS with frontotemporal lobe dementia (ALS-FTD),
Recent Studies
1. Graph theory based MRI analysis of ALS patients
Study
Frontal and temporal white matter (WM) network damage in ALS-FTD patients found using graph theory
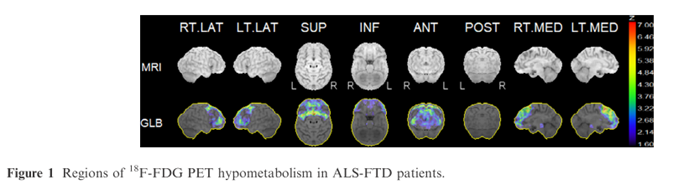
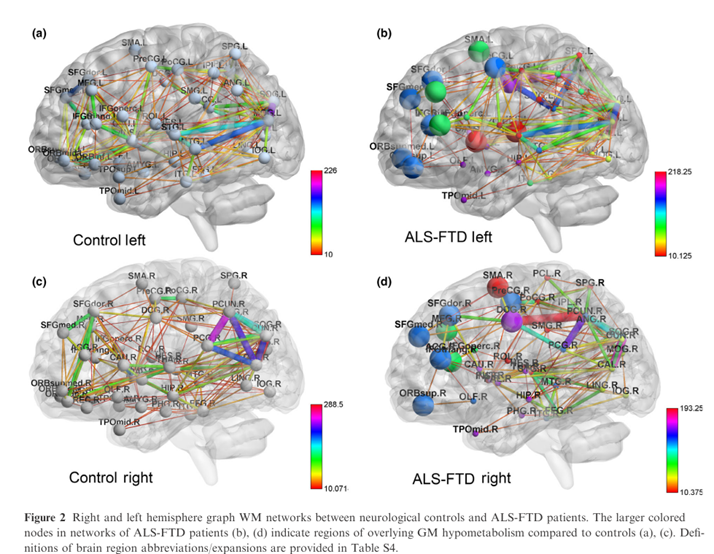
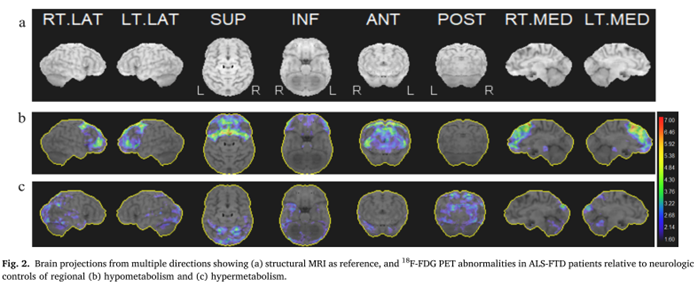
Graph theory network analysis provides brain MRI evidence of a partial continuum of neurodegeneration in patients with UMN-predominant ALS and ALS-FTD 
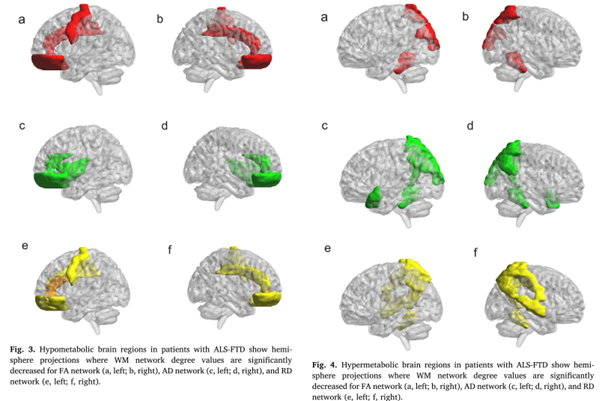
Differing patterns of cortical gray matter pathology identified by multifractal analysis in UMN predominant ALS phenotypes 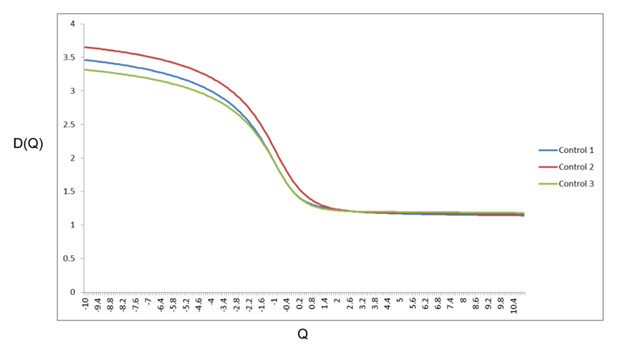
Ongoing Studies in ALS
Diffusion Network model of disease propagation in ALS phenotypes (completed, manuscript in Preparation)
Free water analysis


 An Institute of Eminence
An Institute of Eminence













